Since I migrated my blog from Github Pages to blogdown and Netlify, I wanted to start migrating (most of) my old posts too – and use that opportunity to update them and make sure the code still works. Here I am updating my very first machine learning post: Can we predict flu deaths with Machine Learning and R?. Changes are marked as bold comments.
The main changes I made are:
- using the
tidyversemore consistently throughout the analysis - focusing on comparing multiple imputations from the
micepackage, rather than comparing different algorithms - using
purrr,map(),nest()andunnest()to model and predict the machine learning algorithm over the different imputed datasets
Among the many nice R packages containing data collections is the outbreaks package. It contains a dataset on epidemics and among them is data from the 2013 outbreak of influenza A H7N9 in China as analysed by Kucharski et al. (2014):
- A. Kucharski, H. Mills, A. Pinsent, C. Fraser, M. Van Kerkhove, C. A. Donnelly, and S. Riley. 2014. Distinguishing between reservoir exposure and human-to-human transmission for emerging pathogens using case onset data. PLOS Currents Outbreaks. Mar 7, edition 1. doi: 10.1371/currents.outbreaks.e1473d9bfc99d080ca242139a06c455f.
- A. Kucharski, H. Mills, A. Pinsent, C. Fraser, M. Van Kerkhove, C. A. Donnelly, and S. Riley. 2014. Data from: Distinguishing between reservoir exposure and human-to-human transmission for emerging pathogens using case onset data. Dryad Digital Repository. http://dx.doi.org/10.5061/dryad.2g43n.
I will be using their data as an example to show how to use Machine Learning algorithms for predicting disease outcome.
library(outbreaks)
library(tidyverse)
library(plyr)
library(mice)
library(caret)
library(purrr)
The data
The dataset contains case ID, date of onset, date of hospitalization, date of outcome, gender, age, province and of course outcome: Death or Recovery.
Pre-processing
Change: variable names (i.e. column names) have been renamed, dots have been replaced with underscores, letters are all lower case now.
Change: I am using the tidyverse notation more consistently.
First, I'm doing some preprocessing, including:
- renaming missing data as NA
- adding an ID column
- setting column types
- gathering date columns
- changing factor names of dates (to make them look nicer in plots) and of
province(to combine provinces with few cases)
fluH7N9_china_2013$age[which(fluH7N9_china_2013$age == "?")] <- NA
fluH7N9_china_2013_gather <- fluH7N9_china_2013 %>%
mutate(case_id = paste("case", case_id, sep = "_"),
age = as.numeric(age)) %>%
gather(Group, Date, date_of_onset:date_of_outcome) %>%
mutate(Group = as.factor(mapvalues(Group, from = c("date_of_onset", "date_of_hospitalisation", "date_of_outcome"),
to = c("date of onset", "date of hospitalisation", "date of outcome"))),
province = mapvalues(province, from = c("Anhui", "Beijing", "Fujian", "Guangdong", "Hebei", "Henan", "Hunan", "Jiangxi", "Shandong", "Taiwan"), to = rep("Other", 10)))
I'm also
- adding a third gender level for unknown gender
levels(fluH7N9_china_2013_gather$gender) <- c(levels(fluH7N9_china_2013_gather$gender), "unknown")
fluH7N9_china_2013_gather$gender[is.na(fluH7N9_china_2013_gather$gender)] <- "unknown"
head(fluH7N9_china_2013_gather)
## case_id outcome gender age province Group Date
## 1 case_1 Death m 58 Shanghai date of onset 2013-02-19
## 2 case_2 Death m 7 Shanghai date of onset 2013-02-27
## 3 case_3 Death f 11 Other date of onset 2013-03-09
## 4 case_4 <NA> f 18 Jiangsu date of onset 2013-03-19
## 5 case_5 Recover f 20 Jiangsu date of onset 2013-03-19
## 6 case_6 Death f 9 Jiangsu date of onset 2013-03-21
For plotting, I am defining a custom ggplot2 theme:
my_theme <- function(base_size = 12, base_family = "sans"){
theme_minimal(base_size = base_size, base_family = base_family) +
theme(
axis.text = element_text(size = 12),
axis.text.x = element_text(angle = 45, vjust = 0.5, hjust = 0.5),
axis.title = element_text(size = 14),
panel.grid.major = element_line(color = "grey"),
panel.grid.minor = element_blank(),
panel.background = element_rect(fill = "aliceblue"),
strip.background = element_rect(fill = "lightgrey", color = "grey", size = 1),
strip.text = element_text(face = "bold", size = 12, color = "black"),
legend.position = "bottom",
legend.justification = "top",
legend.box = "horizontal",
legend.box.background = element_rect(colour = "grey50"),
legend.background = element_blank(),
panel.border = element_rect(color = "grey", fill = NA, size = 0.5)
)
}
And use that theme to visualize the data:
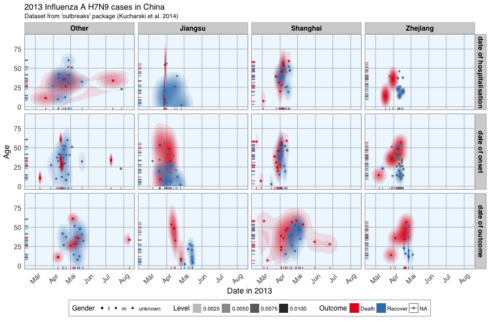
ggplot(data = fluH7N9_china_2013_gather, aes(x = Date, y = age, fill = outcome)) +
stat_density2d(aes(alpha = ..level..), geom = "polygon") +
geom_jitter(aes(color = outcome, shape = gender), size = 1.5) +
geom_rug(aes(color = outcome)) +
scale_y_continuous(limits = c(0, 90)) +
labs(
fill = "Outcome",
color = "Outcome",
alpha = "Level",
shape = "Gender",
x = "Date in 2013",
y = "Age",
title = "2013 Influenza A H7N9 cases in China",
subtitle = "Dataset from 'outbreaks' package (Kucharski et al. 2014)",
caption = ""
) +
facet_grid(Group ~ province) +
my_theme() +
scale_shape_manual(values = c(15, 16, 17)) +
scale_color_brewer(palette="Set1", na.value = "grey50") +
scale_fill_brewer(palette="Set1")
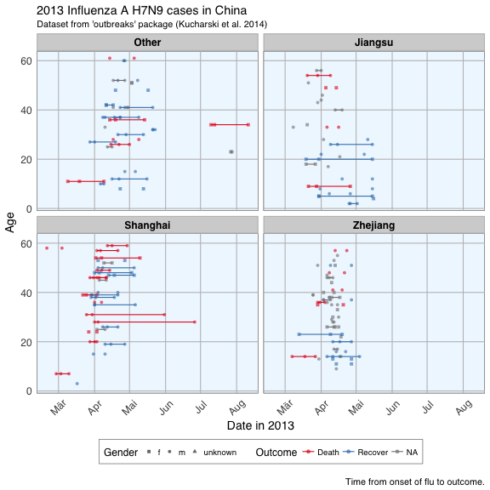
ggplot(data = fluH7N9_china_2013_gather, aes(x = Date, y = age, color = outcome)) +
geom_point(aes(color = outcome, shape = gender), size = 1.5, alpha = 0.6) +
geom_path(aes(group = case_id)) +
facet_wrap( ~ province, ncol = 2) +
my_theme() +
scale_shape_manual(values = c(15, 16, 17)) +
scale_color_brewer(palette="Set1", na.value = "grey50") +
scale_fill_brewer(palette="Set1") +
labs(
color = "Outcome",
shape = "Gender",
x = "Date in 2013",
y = "Age",
title = "2013 Influenza A H7N9 cases in China",
subtitle = "Dataset from 'outbreaks' package (Kucharski et al. 2014)",
caption = "\nTime from onset of flu to outcome."
)
Features
In machine learning-speak features are what we call the variables used for model training. Using the right features dramatically influences the accuracy and success of your model. For this example, I am keeping age, but I am also generating new features from the date information and converting gender and province into numerical values.
dataset <- fluH7N9_china_2013 %>%
mutate(hospital = as.factor(ifelse(is.na(date_of_hospitalisation), 0, 1)),
gender_f = as.factor(ifelse(gender == "f", 1, 0)),
province_Jiangsu = as.factor(ifelse(province == "Jiangsu", 1, 0)),
province_Shanghai = as.factor(ifelse(province == "Shanghai", 1, 0)),
province_Zhejiang = as.factor(ifelse(province == "Zhejiang", 1, 0)),
province_other = as.factor(ifelse(province == "Zhejiang" | province == "Jiangsu" | province == "Shanghai", 0, 1)),
days_onset_to_outcome = as.numeric(as.character(gsub(" days", "",
as.Date(as.character(date_of_outcome), format = "%Y-%m-%d") -
as.Date(as.character(date_of_onset), format = "%Y-%m-%d")))),
days_onset_to_hospital = as.numeric(as.character(gsub(" days", "",
as.Date(as.character(date_of_hospitalisation), format = "%Y-%m-%d") -
as.Date(as.character(date_of_onset), format = "%Y-%m-%d")))),
age = age,
early_onset = as.factor(ifelse(date_of_onset < summary(fluH7N9_china_2013$date_of_onset)[[3]], 1, 0)),
early_outcome = as.factor(ifelse(date_of_outcome < summary(fluH7N9_china_2013$date_of_outcome)[[3]], 1, 0))) %>%
subset(select = -c(2:4, 6, 8))
rownames(dataset) <- dataset$case_id
dataset[, -2] <- as.numeric(as.matrix(dataset[, -2]))
head(dataset)
## case_id outcome age hospital gender_f province_Jiangsu province_Shanghai
## 1 1 Death 87 0 0 0 1
## 2 2 Death 27 1 0 0 1
## 3 3 Death 35 1 1 0 0
## 4 4 <NA> 45 1 1 1 0
## 5 5 Recover 48 1 1 1 0
## 6 6 Death 32 1 1 1 0
## province_Zhejiang province_other days_onset_to_outcome
## 1 0 0 13
## 2 0 0 11
## 3 0 1 31
## 4 0 0 NA
## 5 0 0 57
## 6 0 0 36
## days_onset_to_hospital early_onset early_outcome
## 1 NA 1 1
## 2 4 1 1
## 3 10 1 1
## 4 8 1 NA
## 5 11 1 0
## 6 7 1 1
summary(dataset$outcome)
## Death Recover NA's
## 32 47 57
Imputing missing values
I am using the mice package for imputing missing values. DataScience+ has also other tutorials how to impute data with MICE.
Note: Since publishing this blog post I learned that the idea behind using mice is to compare different imputations to see how stable they are, instead of picking one imputed set as fixed for the remainder of the analysis. Therefore, I changed the focus of this post a little bit: in the old post I compared many different algorithms and their outcome; in this updated version I am only showing the Random Forest algorithm and focus on comparing the different imputed datasets. I am ignoring feature importance and feature plots because nothing changed compared to the old post.
md.pattern()shows the pattern of missingness in the data:
md.pattern(dataset)
## case_id hospital province_Jiangsu province_Shanghai province_Zhejiang
## 42 1 1 1 1 1
## 27 1 1 1 1 1
## 2 1 1 1 1 1
## 2 1 1 1 1 1
## 18 1 1 1 1 1
## 1 1 1 1 1 1
## 36 1 1 1 1 1
## 3 1 1 1 1 1
## 3 1 1 1 1 1
## 2 1 1 1 1 1
## 0 0 0 0 0
## province_other age gender_f early_onset outcome early_outcome
## 42 1 1 1 1 1 1
## 27 1 1 1 1 1 1
## 2 1 1 1 1 1 0
## 2 1 1 1 0 1 1
## 18 1 1 1 1 0 0
## 1 1 1 1 1 1 0
## 36 1 1 1 1 0 0
## 3 1 1 1 0 1 0
## 3 1 1 1 0 0 0
## 2 1 0 0 0 1 0
## 0 2 2 10 57 65
## days_onset_to_outcome days_onset_to_hospital
## 42 1 1 0
## 27 1 0 1
## 2 0 1 2
## 2 0 0 3
## 18 0 1 3
## 1 0 0 3
## 36 0 0 4
## 3 0 0 4
## 3 0 0 5
## 2 0 0 6
## 67 74 277
mice()generates the imputations
dataset_impute <- mice(data = dataset[, -2], print = FALSE)
- by default,
mice()calculates five (m = 5) imputed data sets - we can combine them all in one output with the
complete("long")function - I did not want to impute missing values in the
outcomecolumn, so I have to merge it back in with the imputed data
datasets_complete <- right_join(dataset[, c(1, 2)],
complete(dataset_impute, "long"),
by = "case_id") %>%
select(-.id)
head(datasets_complete)
## case_id outcome .imp age hospital gender_f province_Jiangsu
## 1 1 Death 1 87 0 0 0
## 2 2 Death 1 27 1 0 0
## 3 3 Death 1 35 1 1 0
## 4 4 <NA> 1 45 1 1 1
## 5 5 Recover 1 48 1 1 1
## 6 6 Death 1 32 1 1 1
## province_Shanghai province_Zhejiang province_other days_onset_to_outcome
## 1 1 0 0 13
## 2 1 0 0 11
## 3 0 0 1 31
## 4 0 0 0 20
## 5 0 0 0 57
## 6 0 0 0 36
## days_onset_to_hospital early_onset early_outcome
## 1 5 1 1
## 2 4 1 1
## 3 10 1 1
## 4 8 1 1
## 5 11 1 0
## 6 7 1 1
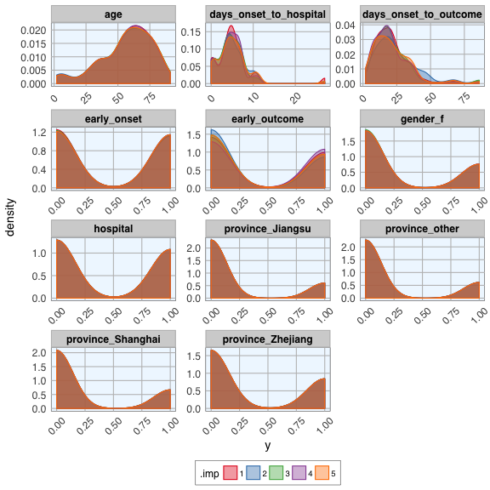
Let's compare the distributions of the five different imputed datasets:
datasets_complete %>%
gather(x, y, age:early_outcome) %>%
ggplot(aes(x = y, fill = .imp, color = .imp)) +
facet_wrap(~ x, ncol = 3, scales = "free") +
geom_density(alpha = 0.4) +
scale_fill_brewer(palette="Set1", na.value = "grey50") +
scale_color_brewer(palette="Set1", na.value = "grey50") +
my_theme()
Test, train and validation data sets
Now, we can go ahead with machine learning!
The dataset contains a few missing values in the outcome column; those will be the test set used for final predictions (see the old blog post for this).
train_index <- which(is.na(datasets_complete$outcome))
train_data <- datasets_complete[-train_index, ]
test_data <- datasets_complete[train_index, -2]
The remainder of the data will be used for modeling. Here, I am splitting the data into 70% training and 30% test data.
Because I want to model each imputed dataset separately, I am using the nest() and map() functions.
set.seed(42)
val_data <- train_data %>%
group_by(.imp) %>%
nest() %>%
mutate(val_index = map(data, ~ createDataPartition(.$outcome, p = 0.7, list = FALSE)),
val_train_data = map2(data, val_index, ~ .x[.y, ]),
val_test_data = map2(data, val_index, ~ .x[-.y, ]))
Machine Learning algorithms
Random Forest
To make the code tidier, I am first defining the modeling function with the parameters I want.
model_function <- function(df) {
caret::train(outcome ~ .,
data = df,
method = "rf",
preProcess = c("scale", "center"),
trControl = trainControl(method = "repeatedcv", number = 5, repeats = 3, verboseIter = FALSE))
}
Next, I am using the nested tibble from before to map() the model function, predict the outcome and calculate confusion matrices.
set.seed(42)
val_data_model <- val_data %>%
mutate(model = map(val_train_data, ~ model_function(.x)),
predict = map2(model, val_test_data, ~ data.frame(prediction = predict(.x, .y[, -2]))),
predict_prob = map2(model, val_test_data, ~ data.frame(outcome = .y[, 2],
prediction = predict(.x, .y[, -2], type = "prob"))),
confusion_matrix = map2(val_test_data, predict, ~ confusionMatrix(.x$outcome, .y$prediction)),
confusion_matrix_tbl = map(confusion_matrix, ~ as.tibble(.x$table)))
Comparing accuracy of models
To compare how the different imputations did, I am plotting
- the confusion matrices:
val_data_model %>%
unnest(confusion_matrix_tbl) %>%
ggplot(aes(x = Prediction, y = Reference, fill = n)) +
facet_wrap(~ .imp, ncol = 5, scales = "free") +
geom_tile() +
my_theme()
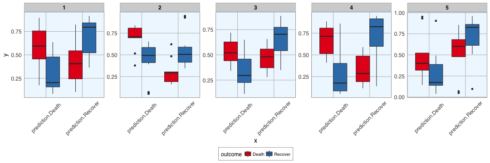
- and the prediction probabilities for correct and wrong predictions:
val_data_model %>%
unnest(predict_prob) %>%
gather(x, y, prediction.Death:prediction.Recover) %>%
ggplot(aes(x = x, y = y, fill = outcome)) +
facet_wrap(~ .imp, ncol = 5, scales = "free") +
geom_boxplot() +
scale_fill_brewer(palette="Set1", na.value = "grey50") +
my_theme()
Hope, you found that example interesting and helpful!
sessionInfo()
## R version 3.5.0 (2018-04-23)
## Platform: x86_64-apple-darwin15.6.0 (64-bit)
## Running under: macOS High Sierra 10.13.4
##
## Matrix products: default
## BLAS: /System/Library/Frameworks/Accelerate.framework/Versions/A/Frameworks/vecLib.framework/Versions/A/libBLAS.dylib
## LAPACK: /Library/Frameworks/R.framework/Versions/3.5/Resources/lib/libRlapack.dylib
##
## locale:
## [1] de_DE.UTF-8/de_DE.UTF-8/de_DE.UTF-8/C/de_DE.UTF-8/de_DE.UTF-8
##
## attached base packages:
## [1] stats graphics grDevices utils datasets methods base
##
## other attached packages:
## [1] bindrcpp_0.2.2 knitr_1.20 RWordPress_0.2-3 caret_6.0-79
## [5] mice_2.46.0 lattice_0.20-35 plyr_1.8.4 forcats_0.3.0
## [9] stringr_1.3.1 dplyr_0.7.4 purrr_0.2.4 readr_1.1.1
## [13] tidyr_0.8.0 tibble_1.4.2 ggplot2_2.2.1 tidyverse_1.2.1
## [17] outbreaks_1.3.0
##
## loaded via a namespace (and not attached):
## [1] nlme_3.1-137 bitops_1.0-6 lubridate_1.7.4
## [4] RColorBrewer_1.1-2 dimRed_0.1.0 httr_1.3.1
## [7] rprojroot_1.3-2 tools_3.5.0 backports_1.1.2
## [10] R6_2.2.2 rpart_4.1-13 lazyeval_0.2.1
## [13] colorspace_1.3-2 nnet_7.3-12 withr_2.1.2
## [16] tidyselect_0.2.4 mnormt_1.5-5 compiler_3.5.0
## [19] cli_1.0.0 rvest_0.3.2 xml2_1.2.0
## [22] labeling_0.3 bookdown_0.7 scales_0.5.0
## [25] sfsmisc_1.1-2 DEoptimR_1.0-8 psych_1.8.4
## [28] robustbase_0.93-0 randomForest_4.6-14 digest_0.6.15
## [31] foreign_0.8-70 rmarkdown_1.9 pkgconfig_2.0.1
## [34] htmltools_0.3.6 highr_0.6 rlang_0.2.0
## [37] readxl_1.1.0 ddalpha_1.3.3 rstudioapi_0.7
## [40] XMLRPC_0.3-0 bindr_0.1.1 jsonlite_1.5
## [43] ModelMetrics_1.1.0 RCurl_1.95-4.10 magrittr_1.5
## [46] Matrix_1.2-14 Rcpp_0.12.16 munsell_0.4.3
## [49] abind_1.4-5 stringi_1.2.2 yaml_2.1.19
## [52] MASS_7.3-50 recipes_0.1.2 grid_3.5.0
## [55] parallel_3.5.0 crayon_1.3.4 haven_1.1.1
## [58] splines_3.5.0 hms_0.4.2 pillar_1.2.2
## [61] reshape2_1.4.3 codetools_0.2-15 stats4_3.5.0
## [64] CVST_0.2-1 magic_1.5-8 XML_3.98-1.11
## [67] glue_1.2.0 evaluate_0.10.1 blogdown_0.6
## [70] modelr_0.1.2 foreach_1.4.4 cellranger_1.1.0
## [73] gtable_0.2.0 kernlab_0.9-26 assertthat_0.2.0
## [76] DRR_0.0.3 xfun_0.1 gower_0.1.2
## [79] prodlim_2018.04.18 broom_0.4.4 e1071_1.6-8
## [82] class_7.3-14 survival_2.42-3 geometry_0.3-6
## [85] timeDate_3043.102 RcppRoll_0.2.2 iterators_1.0.9
## [88] lava_1.6.1 ipred_0.9-6