This post is a continuation of my previous tutorial regarding the visualization of National Health and Nutrition Examination Survey (NHANES). Both posts aim to show how we can use the functions of ggplot to visualize the data. Also, I show the steps I follow to make data ready for visualization..
NHANES is a survey which combines interviews, physical examinations, and laboratory testing of Americans. You can find information about NHANES at the program’s website.
As I mentioned in the other post, no conclusion should be made from the plots or results, as this post is only for illustration of ggplot functions.
In this post, I will plot the levels of plasma lipids across age and gender. Most known blood lipids include LDL cholesterol, HDL cholesterol, and triglycerides. Those lipids are measured in NHANES survey and I will explore their relationship with age and gender. Higher blood lipids (LDL cholesterol and triglycerides) is associated with increased risk of cardiovascular disease. In contrary, higher levels of HDL cholesterol is related to lower risk of cardiovascular disease.
Ok, now lets dive into NHANES data.
Libraries and data
First I will load the neccessary libraries
library(tidyverse) library(RNHANES) library(ggsci) library(ggthemes)
Load and merge the data in one dataset.
data1 %>%
left_join(nhanes_load_data("TRIGLY_H", "2013-2014"), by="SEQN") %>%
left_join(nhanes_load_data("DEMO_H", "2013-2014"), by="SEQN")
Checking the data
Creating the dataset with variables of interest
data2 = data1 %>% select(SEQN, RIAGENDR, RIDAGEYR, LBDHDD, LBDLDL, LBXTR)
-
SEQN: id
RIAGENDR: gender
RIDAGEYR: age in years
LBDHDD: HDL-cholesterol
LBDLDL: LDL-cholesterol
LBXTR: triglyceride
Summary of the data:
summary(data2)
SEQN RIAGENDR RIDAGEYR LBDHDD LBDLDL LBXTR
Min. :73557 Min. :1.000 Min. : 6.00 Min. : 10.00 Min. : 14.0 Min. : 13.0
1st Qu.:76132 1st Qu.:1.000 1st Qu.:16.00 1st Qu.: 42.00 1st Qu.: 81.0 1st Qu.: 60.0
Median :78695 Median :2.000 Median :35.00 Median : 51.00 Median :103.0 Median : 88.0
Mean :78676 Mean :1.513 Mean :37.04 Mean : 53.11 Mean :106.2 Mean : 112.3
3rd Qu.:81218 3rd Qu.:2.000 3rd Qu.:56.00 3rd Qu.: 61.00 3rd Qu.:127.0 3rd Qu.: 133.0
Max. :83731 Max. :2.000 Max. :80.00 Max. :173.00 Max. :375.0 Max. :4233.0
NA's :667 NA's :5186 NA's :5145
I will remove missing values and exclude participants younger than 40 years. I will also create a new variable such as age-groups and recode gender variable as “Men” and “Women.” I use the function case_when to create the age-group variable. You can do the same with the function ifelse(), but I prefer case_when. To remove missings and those younger than 40 years old I use the function filter.
data3 = data2 %>%
mutate(
gender = recode_factor(RIAGENDR, `1` = "Men", `2` = "Women"),
age_group = case_when(
RIDAGEYR = 40 & RIDAGEYR = 45 & RIDAGEYR = 50 & RIDAGEYR = 55 & RIDAGEYR = 60 & RIDAGEYR = 65 & RIDAGEYR = 70 & RIDAGEYR = 75 & RIDAGEYR %
filter(!is.na(LBDLDL), !is.na(LBXTR), !is.na(LBDHDD), age_group != "NA")
Visualization
Lets take a look at the data:
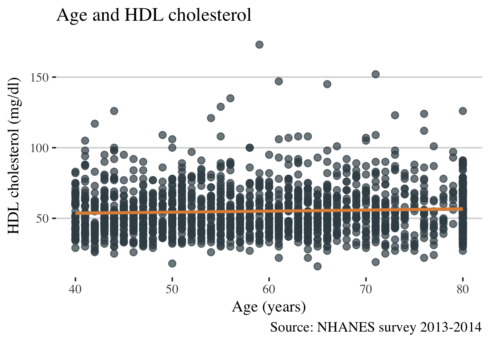
ggplot(data3, aes(RIDAGEYR, LBDHDD)) +
geom_point(alpha = 0.7, size = 2, color = "#3C4D54") +
geom_smooth(size = 1, color = "#E08F44") +
theme_hc() +
theme(text = element_text(family = "serif", size = 11)) +
labs(
title = "Age and HDL cholesterol",
x = "Age (years)",
y = "HDL cholesterol (mg/dl)",
caption = "Source: NHANES survey 2013-2014"
)
This plot shows that levels of HDL cholesterol are similar across the age in NHANES data. Now let see other lipids concerning age.
First I will transform the dataset and after I will create the plot.
long = data3 %>% select(SEQN, gender, RIDAGEYR, LBXTR, LBDLDL, LBDHDD, age_group) %>% gather(lipid, value, LBXTR:LBDLDL:LBDHDD) %>% mutate(lipid = recode(lipid, `LBXTR` = "Triglyceride", `LBDLDL` = "LDL-cholesterol", `LBDHDD` = "HDL-cholesterol" )) head(long) SEQN gender RIDAGEYR age_group lipid value 1 73559 Men 72 70-74 Triglyceride 51 2 73561 Women 73 70-74 Triglyceride 75 3 73564 Women 61 60-64 Triglyceride 64 4 73581 Men 50 50-54 Triglyceride 93 5 73596 Women 57 55-59 Triglyceride 87 6 73607 Men 75 75-80 Triglyceride 139
Plot the lipids across age:
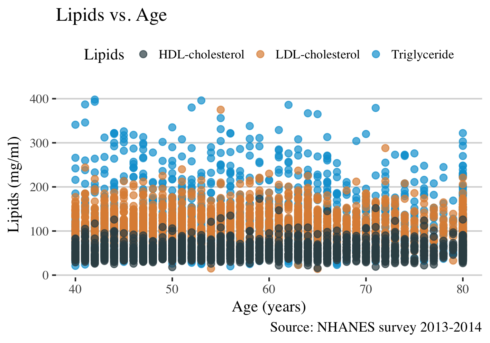
ggplot(long, aes( RIDAGEYR, value, color = lipid)) +
geom_point(alpha = 0.7, size = 2) +
scale_color_jama() +
theme_hc() +
theme(text = element_text(family = "serif", size = 11), legend.position="top") +
xlab("Age (years)") +
ylab("Lipids (mg/ml)") +
ggtitle("Lipids vs. Age") +
labs(
caption = "Source: NHANES survey 2013-2014",
col="Lipids")
Given that the data points are overlapping each other is difficult to make any conclusion. The use of facet_grid function will solve this issue.
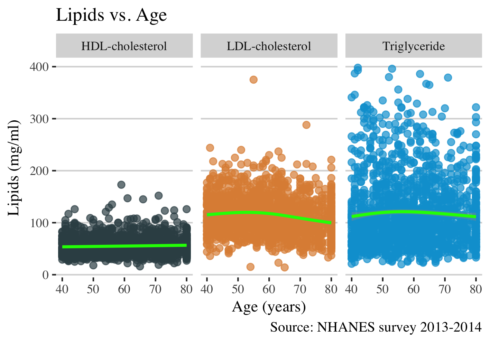
ggplot(long, aes(RIDAGEYR, value, color = lipid)) +
geom_point(alpha = 0.7, size = 2) +
geom_smooth(size = 1, color = "green") +
scale_color_jama() +
facet_grid(~lipid) +
theme_hc() +
theme(text = element_text(family = "serif", size = 11), legend.position="none") +
xlab("Age (years)") +
ylab("Lipids (mg/ml)") +
ggtitle("Lipids vs. Age") +
labs(
caption = "Source: NHANES survey 2013-2014",
col="Lipids")
This plot shows that the levels of LDL cholesterol increases slightly between ages 50-60 and then after decreases. To have a comprehensive view of the relationship between age and lipids levels, I think I need to calculate the mean of lipids per age group and to plot it.
Calculate the mean of lipids by age group.
long_mean = long %>%
group_by(age_group, lipid) %>%
summarise(mean = mean(value))
head(long_mean)
# A tibble: 6 x 3
# Groups: age_group [2]
age_group lipid mean
1 40-44 HDL-cholesterol 54.05242
2 40-44 LDL-cholesterol 114.65726
3 40-44 Triglyceride 110.68145
4 45-49 HDL-cholesterol 51.99517
5 45-49 LDL-cholesterol 120.09662
6 45-49 Triglyceride 121.45894
Dataset looks good and now lets build the plot. I will use geom_point to plot the mean and geom_line to connect the points with line.
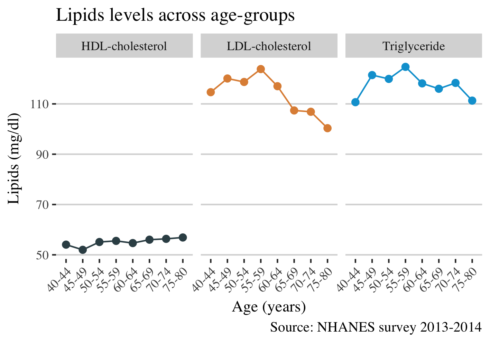
ggplot(long_mean, aes(age_group, mean, color = lipid, group = lipid)) +
geom_point(alpha = 1, size = 2) +
geom_line() +
facet_grid(~lipid) +
scale_color_jama() +
theme_hc() +
theme(
text = element_text(family = "serif", size = 11),
legend.position="none",
axis.text.x = element_text(angle = 45, hjust = 1)
) +
labs(
title = "Lipids levels across age-groups",
x = "Age (years)",
y = "Lipids (mg/dl)",
caption = "Source: NHANES survey 2013-2014",
col="")
It is visiable that the levels of LDL cholesterol are decreasing with advancing age. We suspected a decrease from the plot above, but here is more noticeable. The reason for this decline may be attributed to medication of dyslipidemia.
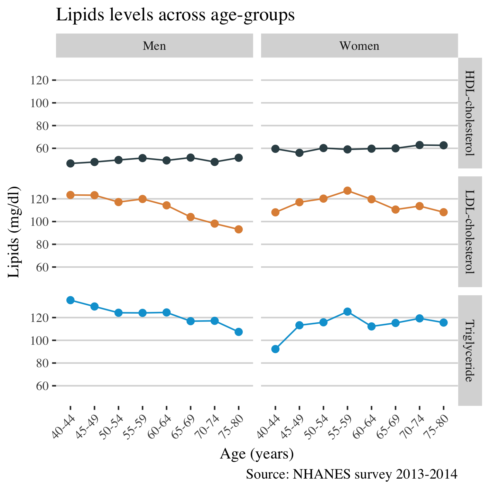
And finally, I am interested to see whether there are gender differences in lipids levels across age.
long %>%
group_by(age_group, lipid, gender) %>%
summarise(mean = mean(value)) %>%
ggplot(aes(age_group, mean, color = lipid, group = lipid)) +
geom_point(alpha = 1, size = 2) +
geom_line() +
facet_grid(lipid~gender) +
scale_color_jama() +
theme_hc() +
theme(
text = element_text(family = "serif", size = 11),
legend.position="none",
axis.text.x = element_text(angle = 45, hjust = 1)
) +
labs(
title = "Lipids levels across age-groups",
x = "Age (years)",
y = "Lipids (mg/dl)",
caption = "Source: NHANES survey 2013-2014",
col="")
Conclusion
I found that the levels of LDL cholesterol and triglycerides decrease after the age of 55 years, probably due to medication. Men experience a larger decrease in LDL cholesterol than women. Compared to men, women had higher (better) levels of HDL cholesterol. Calculating the overall mean of lipids by age group was a better approach for visualizing the differences of lipid levels in Americans.
Feel free to comment if you have any question regarding the code or plots.